|
||
|
|
||
Trichoderma guizhouense NJAU 4742 Genome Database | ||
|
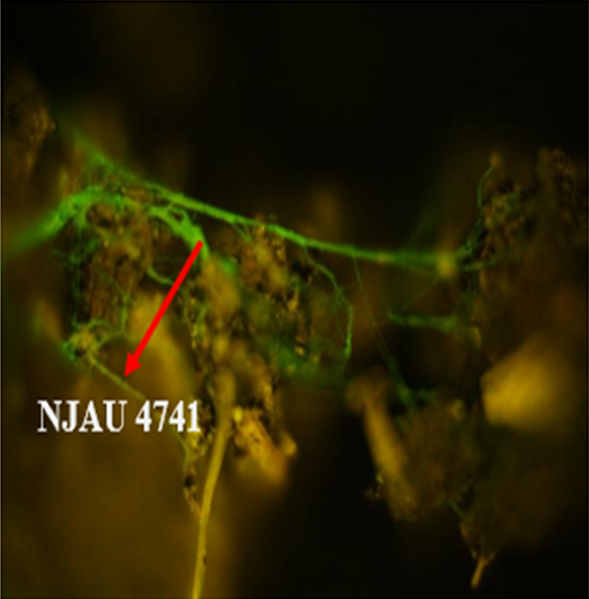
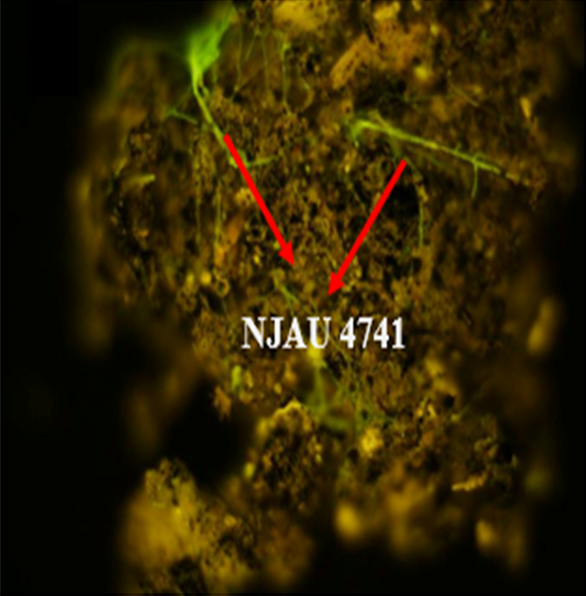
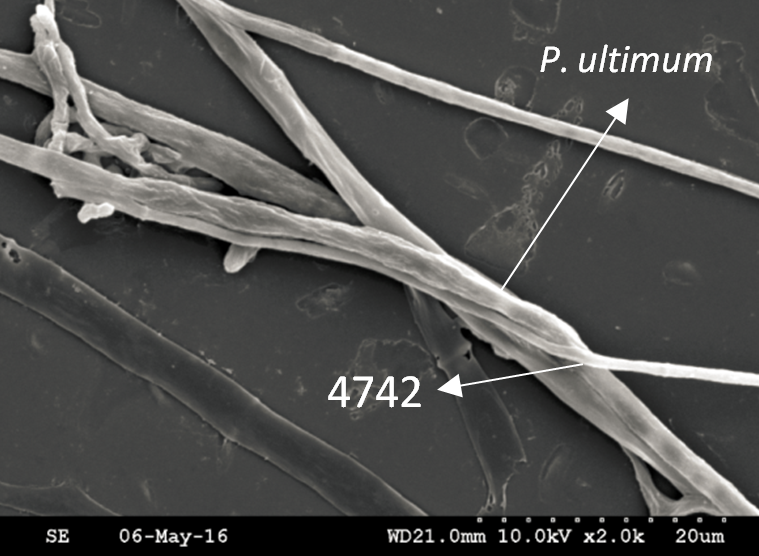
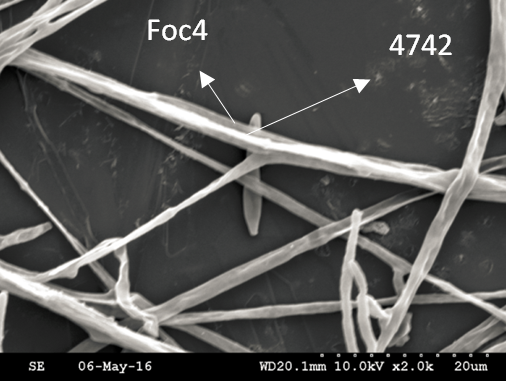
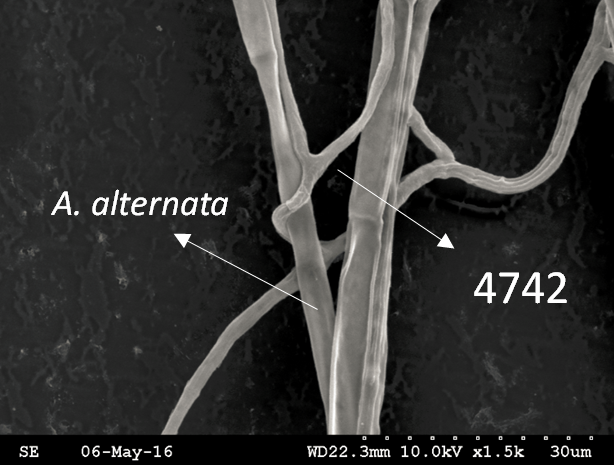
Trichoderma guizhouense NJAU 4742 was isolated from the soil with compost of traditional Chinese medicine in Baguazhou, Nanjing city, Jiangsu province, China. T. guizhouense NJAU 4742 can colonize in plant rhizosphere efficiently, enhance plant growth and suppress various causative agents of plant soil borne diseases, including Fusarium oxysporum, Botrytis cinerea, Sclerotinia sclerotiorum, Rhizoctonia solani and Sclerotium rolfsii, etc. T. guizhouense NJAU 4742 belongs to Harzianum clade but it has more powerful antagonistic ability than T. harzianum CBS 226.95. T. guizhouense NJAU 4742 is an very ideal strain to be used in development of bio-fungicides and improvement of plant health. To understand and make full use of the genetic resource, we sequenced T. guizhouense NJAU 4742 genome sequences using Roche 454 technology, which generated whole genome shotgun (WGS) sequences that were asssembled to obtain the first version of genome sequences. This version of genome assembly contained 416 contigs within 63 scaffold sequences. The N50 lengths of scaffold and contig are 2,4Mb and 181kb, respectively. In the genome assembly, about 1.42% bases were identified as repeats and 296 non-coding RNAs (212 tRNAs, 42rRNAs, 9snRNAs and 33 snoRNAs) and 12,443 protein coding genes were annotated. To facilitate the usage of genomics data, we developed the genome database for T. guizhouense NJAU 4742 (TGNGDB) which is a highly intergrated information system for data storage, retrieval, visualization and analysis. The database provides an integrated representation of large-scale, genome-wide sequence assembly, repeats, nocoding RNAs, protein-coding genes and functional annotation including domain and family, Gene Ontology and pathway. Besides, in order to promote comparative genomic research, the orthologue analysis was performed based on six Trichoderma species and user can conveniently obtain homology genes shared by different species.
Citations: If you wish to use our TGNGDB, please refer to our publication. |
||