4.2 Task setting
4.2.1 Retrieve public samples for re-analysis
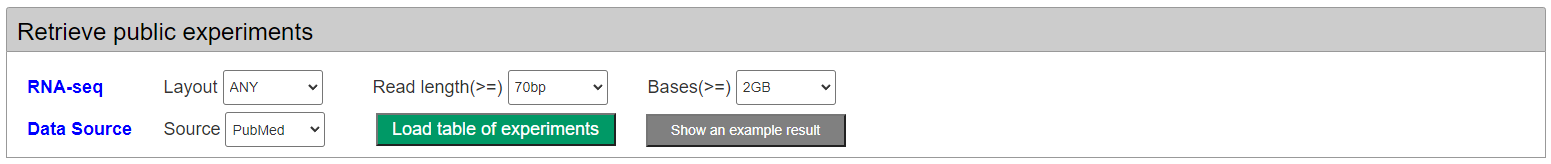
Firstly, users can retrieve the public RNA-seq samples from the platform database by multiple conditions, such as RNA-seq read length, layout, volumen and datasource.
4.2.2 Set groups
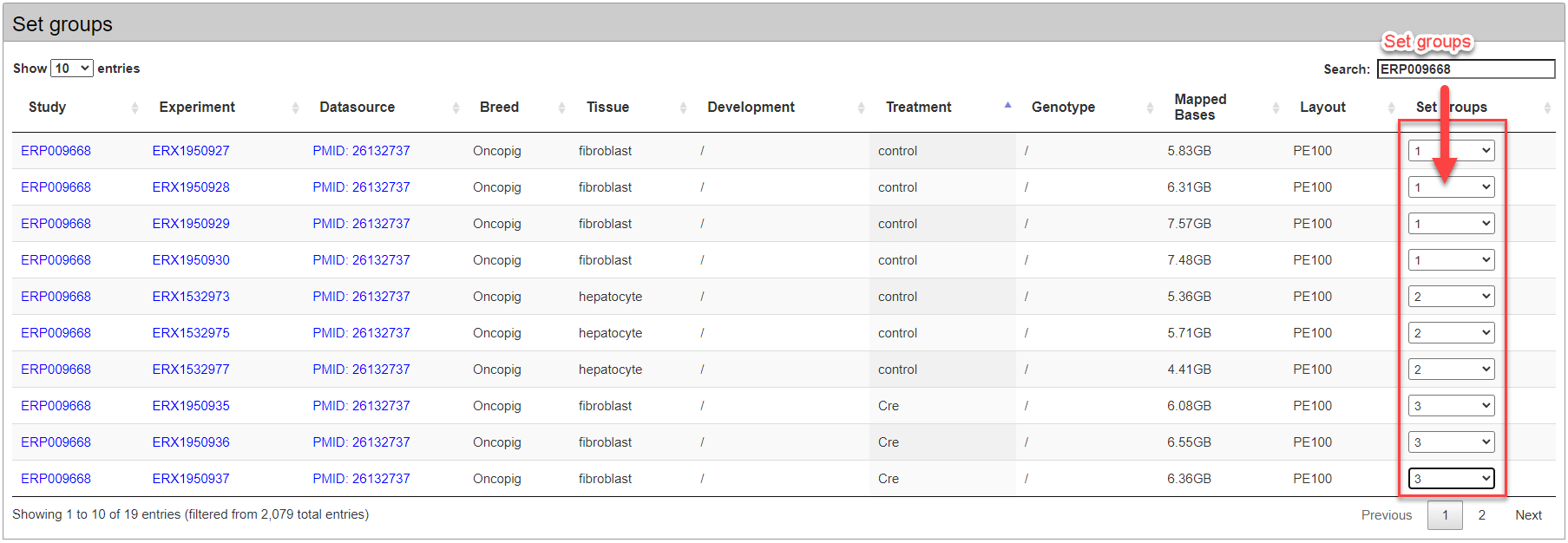
In the retrieved public samples, users should set at least 4 experimental groups by breed, genotype, tissue, development and treatment.
4.2.3 upload samples for co-analysis
To perform co-analysis with public RNA-seq samples, users can upload their RNA-seq data through the following interface. Refer to the
"co-analysis" page to see how to prepare user's own RNA-seq data.
4.2.4 Set group labels
In addition to the default group labels such as A, B, C, E, ..., users can also customize group label to represent group feature.
4.2.5 set methods and parameters
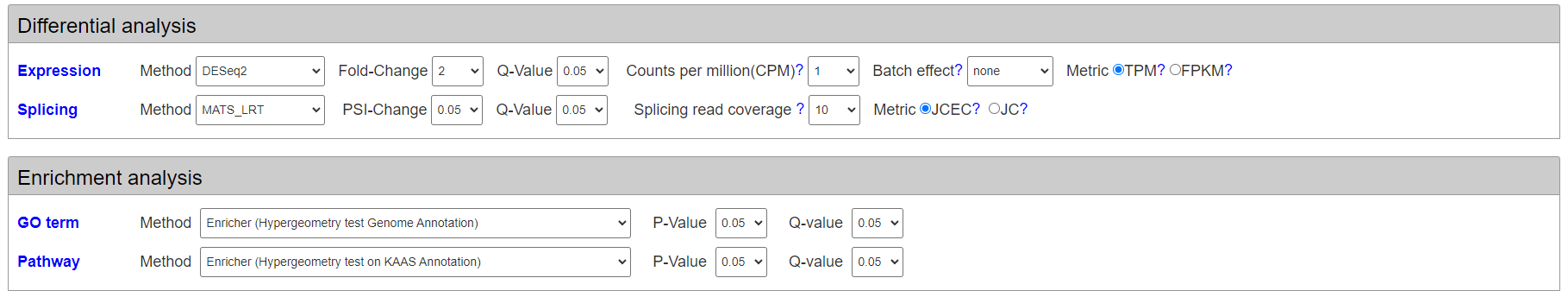
After setting groups, users can select methods and parameters for differential expression analysis(DESeq2/edgeR) and differential splicing analysis(MATS_LRT/rMATS_paired/rMATS_unpaired) and corresponding parameters(fold changes and pvalues).
Moreover, users can select different methods for enrichment analysis on differentially expressed genes and differentially spliced genes. The enrichment analysis methods include Hyergeometry test and Gene Set Enrichment Analysis. The subject of enrichment analysis include gene ontology and KEGG pathways.
4.2.6 job request
The analysis task must be manually confirmed by email to avoid tasks from non-human submission. Users must offer email address to accept confirmation notice or retrieve analysis results.