4 Specifical expression analysis
4.1 Analysis method
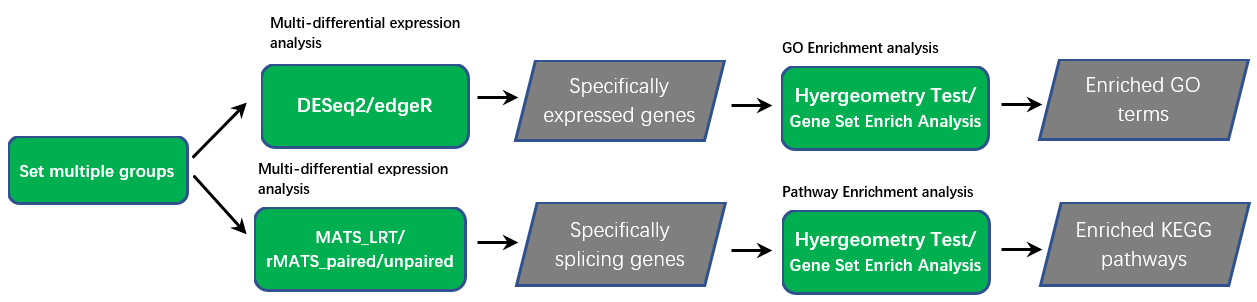
The platform allows users to set multiple groups according to experimental conditions (tissues, developmental stages, stress treatments) to identify specifically expressed and spliced genes. Like differential expression analysis, two commonly used differential expression methods, DESeq2 and edgeR, were also used to compare overall gene expression while statistical models implemented in rMATS were used to compare alternative splicing. The condition specific (e.g. tissue specific) expression or alternative splicing was identified if a gene’s expression or PSI was higher/lower than all other conditions.
When possible (study not completely confounded with treatment), batch effects due to studies were adjusted using DESeq2 or edgeR. The next step is enrichment analysis based on specifically expressed genes and specifically spliced genes.