3.3 Result display
3.3.1 Parameter setting
The first section shows the groups and relative parameters set by users when submit an analysis task. The analysis result files can be downloaded here.
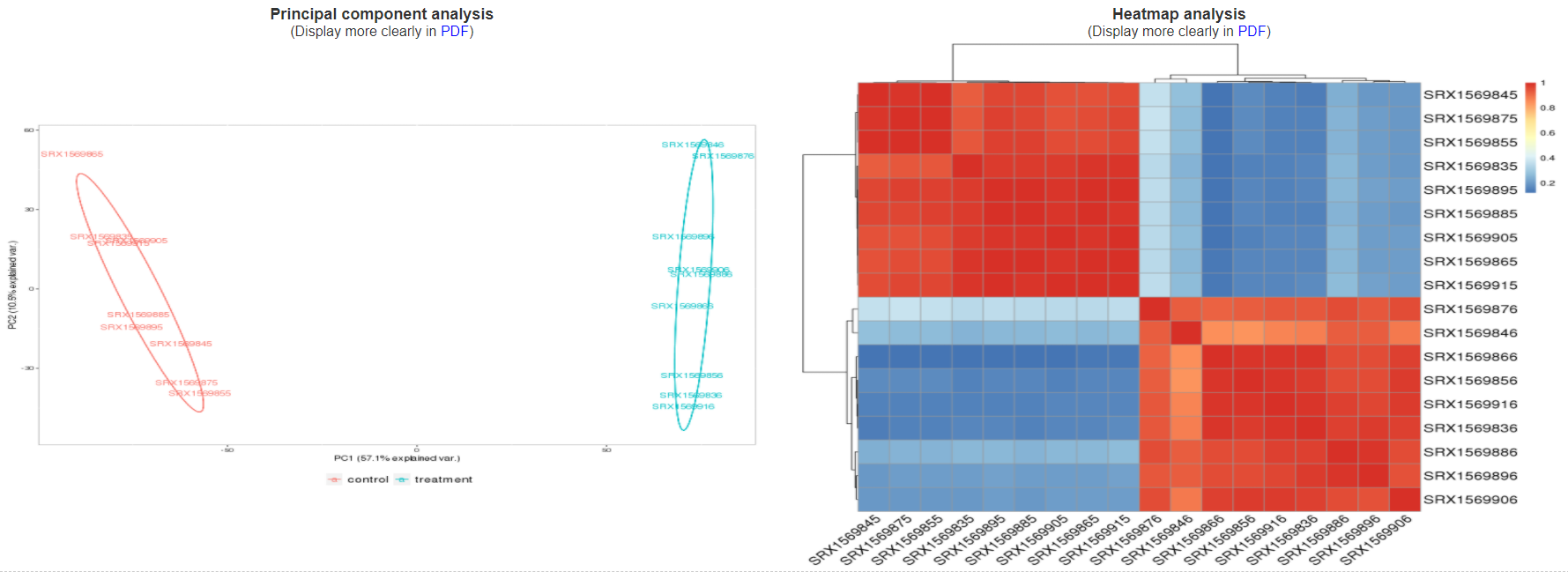
3.3.2 Sample cluster based on expression profiles
This section include two graphics from principal component analysis and heatmap analysis. By the two cluster graphics, users can learn the general affection on gene expression profiles from different experimental conditions.
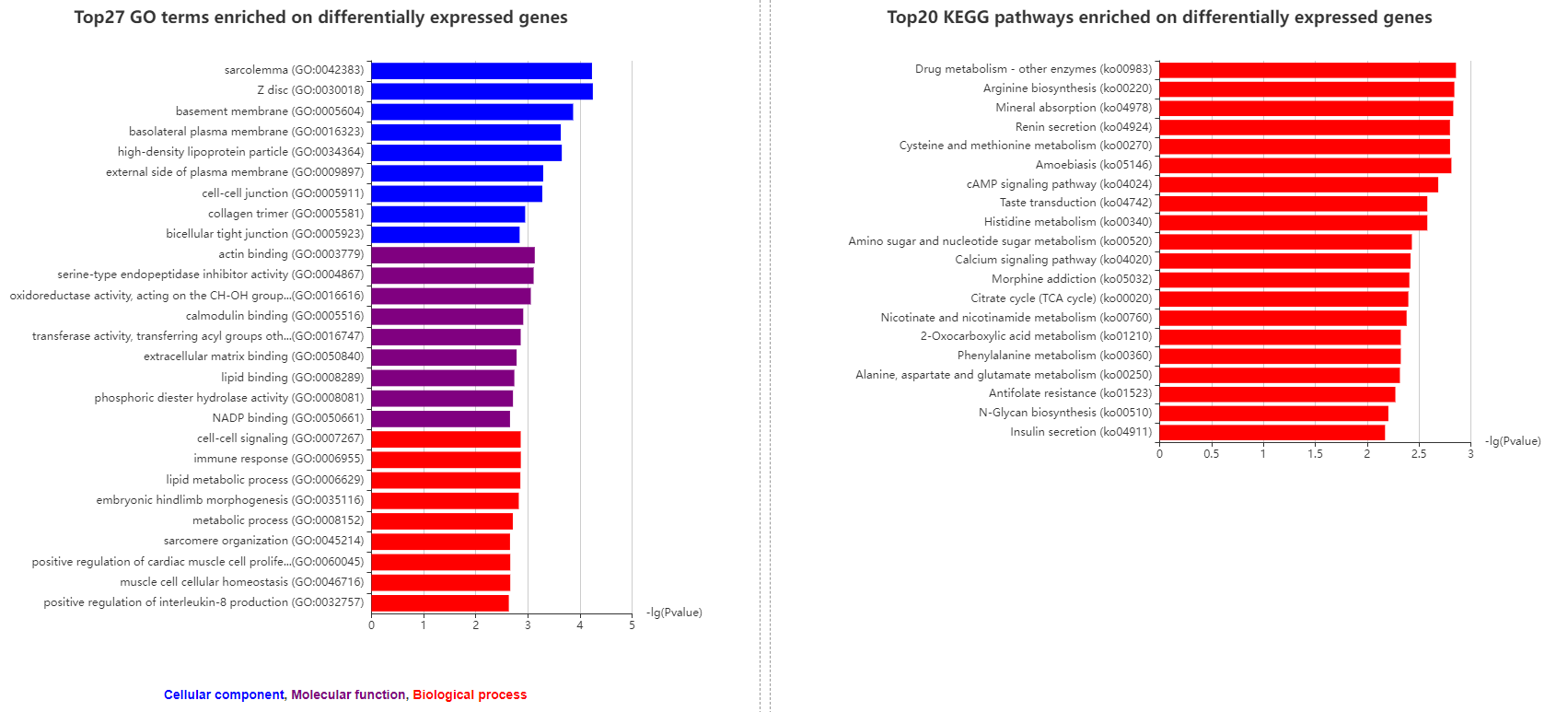
3.3.3 Enriched GO and KEGG pathway terms on differentially expressed genes
Two column graphics are used to show enriched GO and KEGG pathway terms on differentially expressed genes.
3.3.4 List of differentially expressed genes
The differentially expressed genes are listed in an interactive table. Click the "comparison" link to open gene page showing the gene expression level in each sample.
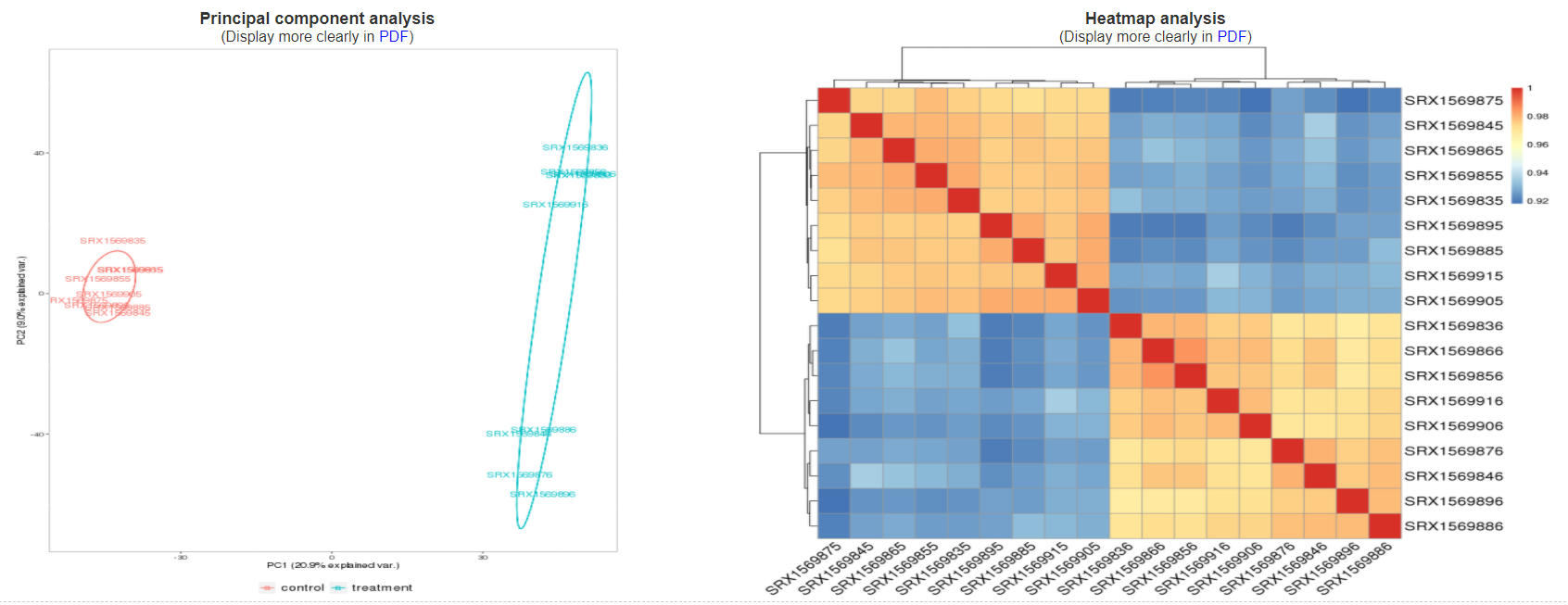
3.3.5 Sample cluster based on alternative splicing profiles
This section include two graphics from principal component analysis and heatmap analysis based on alternative splicing profiles. By the two cluster graphics, users can learn the general affection on gene alternative splicing from different experimental conditions.
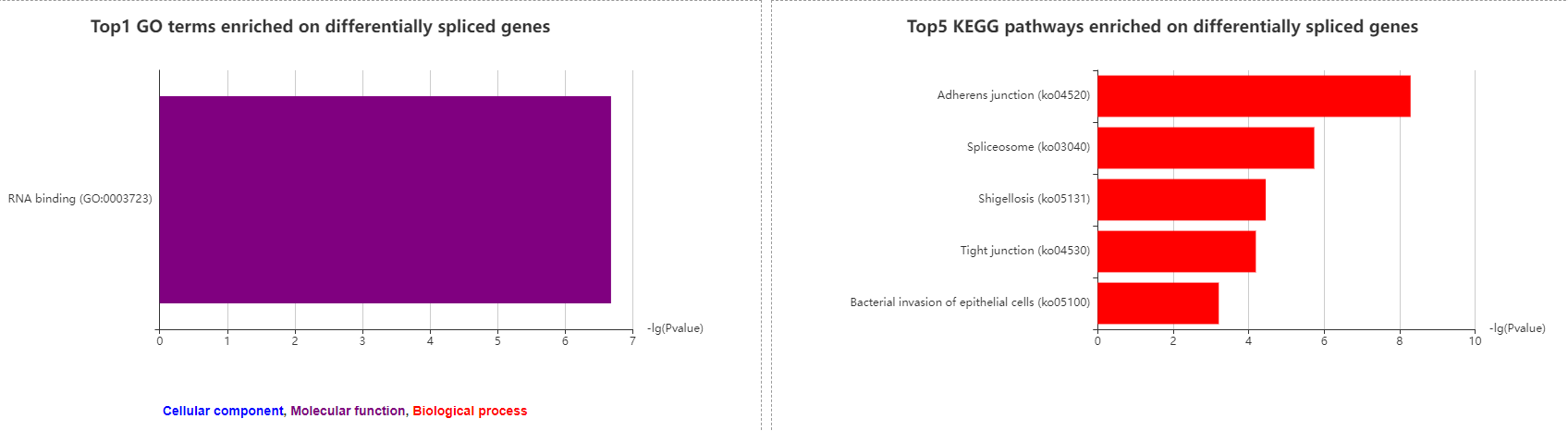
3.3.6 Enriched GO and KEGG pathway terms on differentially spliced genes
Two column graphics are used to show enriched GO and KEGG pathway terms on differentially spliced genes.
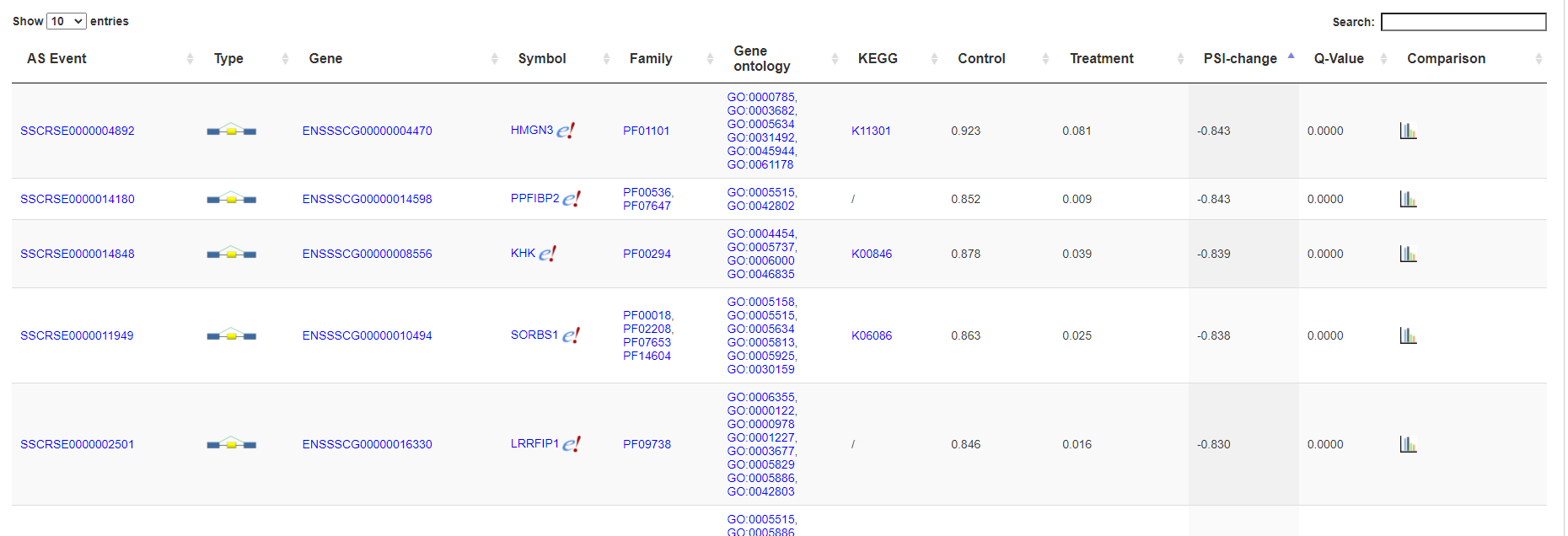
3.3.7 List of differentially spliced genes
The differentially spliced genes are listed in an interactive table. Click the "comparison" link to open alternative splicing page showing the alternative splicing profiles in each sample.